Difference between revisions of "Main Page"
m (Reverted edits by MediaWiki default (talk) to last revision by Andy.jenkinson) |
|||
| (83 intermediate revisions by 21 users not shown) | |||
| Line 1: | Line 1: | ||
<big>'''Welcome To BioDAS.org'''</big> | <big>'''Welcome To BioDAS.org'''</big> | ||
| − | + | ==Notice== | |
| + | |||
| + | <span style="color: darkred;">PLEASE NOTE: </span>a new version of DAS was recently released! See the [[#Versions of DAS|Versions of DAS]] section below for more information. | ||
== About DAS == | == About DAS == | ||
The <b>Distributed Annotation System</b> (DAS) defines a communication protocol used | The <b>Distributed Annotation System</b> (DAS) defines a communication protocol used | ||
| − | to exchange | + | to exchange [http://en.wikipedia.org/wiki/Genome_project#Genome_annotation annotations] on genomic or protein sequences. |
| − | should not be provided by single centralized databases, but instead be spread | + | It is motivated by the idea that such annotations |
| + | should not be provided by single centralized databases, but should instead be spread | ||
over multiple sites. Data distribution, performed by DAS servers, is separated | over multiple sites. Data distribution, performed by DAS servers, is separated | ||
| − | from visualization, which is done by DAS clients. | + | from visualization, which is done by DAS clients. The advantages of this system are that control over the data is retained by data providers, data is freed from the constraints of specific organisations and the normal issues of release cycles, API updates and data duplication are avoided. |
DAS is a client-server system in which a single client integrates information from | DAS is a client-server system in which a single client integrates information from | ||
| − | multiple servers. It allows a single machine to gather up | + | multiple servers. It allows a single machine to gather up sequence annotation |
information from multiple distant web sites, collate the information, and display it | information from multiple distant web sites, collate the information, and display it | ||
to the user in a single view. Little coordination is needed among the various | to the user in a single view. Little coordination is needed among the various | ||
| Line 19: | Line 22: | ||
DAS is heavily used in the genome bioinformatics community. Over the last years we have also | DAS is heavily used in the genome bioinformatics community. Over the last years we have also | ||
seen growing acceptance in the protein sequence and structure communities. | seen growing acceptance in the protein sequence and structure communities. | ||
| − | |||
== What can I do with DAS ? == | == What can I do with DAS ? == | ||
| − | A | + | A DAS-enabled website or application can aggregate complex and high-volume data from external providers in an efficient manner. For the biologist, this means the ability to "plug in" the latest data, possibly including a user's own data. For the application developer, this means protection from data format changes and the ability to add new data with minimal development cost. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Here are some examples of DAS-enabled applications or websites for end users: | |
| − | * | + | * [http://www.biodalliance.org/ Dalliance] Interactive web-based genome viewer |
| + | * [http://www.broadinstitute.org/igv/ IGV] Integrative Genome Viewer java based browser for many genomes | ||
| + | * [http://www.ensembl.org/info/data/ensembl_das.html Ensembl] uses DAS to pull in genomic, gene and protein annotations. It also provides data via DAS. | ||
| + | * [http://www.gmod.org/gbrowse_installation Gbrowse] is a generic genome browser, and is both a consumer and provider of DAS. | ||
| + | * [http://genoviz.sourceforge.net IGB] is a desktop application for viewing genomic data. | ||
| + | * [http://www.efamily.org.uk/software/dasclients/spice/ SPICE] is an application for projecting protein annotations onto 3D structures. | ||
| + | * [http://www.ebi.ac.uk/dasty/ Dasty2] is a web-based viewer for protein annotations | ||
| + | * [http://www.jalview.org Jalview] is a multiple alignment editor. | ||
| + | * [http://biocomp.cnb.csic.es/das/pepper.jsp PeppeR] is a graphical viewer for 3D electron microscopy data. | ||
| + | * [http://www.dasmi.de DASMI] is an integration portal for protein interaction data. | ||
| + | * [http://dasher.sbc.su.se DASher] is a Java-based viewer for protein annotations. | ||
| + | * [http://bioware.ucd.ie/epic/ EpiC] presents structure-function summaries for antibody design. | ||
| + | * [http://www.bioinformatics.org/strap/ STRAP] is a STRucture-based sequence Alignment Program. | ||
| − | + | Hundreds of DAS servers are currently running worldwide, including those provided by the [http://www.ebi.ac.uk/ European Bioinformatics Institute], [http://www.ensembl.org/ Ensembl], the [http://www.sanger.ac.uk/ Sanger Institute], [http://genome.ucsc.edu/ UCSC], [http://www.wormbase.org/ WormBase], [http://www.flybase.org/ FlyBase], [http://www.tigr.org/ TIGR], and [http://www.ebi.ac.uk/uniprot-das/ UniProt]. For a listing of all available DAS sources please visit the [[DasRegistry]]. | |
| − | + | == Versions of DAS == | |
| − | + | The original DAS specification was written by Lincoln Stein, Sean Eddy, and Robin Dowell. It is widely adopted and well supported, particularly throughout Europe, and is the basis for a large number of existing clients and servers. The protocol has been developed incrementally since its inception and thus there are several successive versions of the specification, each expanding on the last whilst retaining a focus on backwards compatibility. The current specification of DAS is version [[DAS1.6|1.6]]. | |
| − | + | Though mature, the protocol continues to be extended to cater for the needs of the DAS community via extensions to the specification. Together, these extensions form an "extended specification". The current version is [[DAS1.6E|1.6E]]. | |
| − | + | <b>NOTE:</b> The DAS/2 project is an entirely separate specification which although based on the DAS architecture is not backwards compatible with existing servers and clients. It is described separately on the [[DAS/2]] page. | |
| − | [[DAS | + | The various specification versions on this site are described [[DAS specification|here]]. |
| − | + | <br> | |
| + | '''Evolution''' | ||
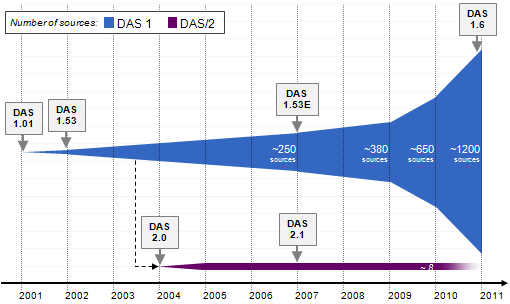
| − | + | [[Image:DasEvolution2.PNG]] | |
| − | + | Graphic representation of the evolution of [http://www.biotnet.org/training-materials/versions-das "Versions of DAS"] over the years. The different colors identify the version/specification and the white numbers an estimation of available sources per year. | |
| − | == | + | == Further Reading == |
| − | + | If you would like to find out more, see a description of the [[DAS/1|DAS protocol]]. Additional information is also available in the form of an [[DasFAQ|FAQ]] page, and a more extensive reference: [[Everything DAS]]. There is also a list of DAS related [[DAS_publications|publications]]. | |
| − | |||
| + | The [[BioDAS:Community_Portal|Community Portal]] page contains information on the various DAS mailing lists. | ||
| − | + | DAS is often represented in workshops, training courses and conferences. A list of past and future events is available on the [[Current events]] page. | |
| + | == Contribute to this Wiki == | ||
Want to contribute to the Wiki? please see some beginners instructions at the [[WikiGetStarted]] page. | Want to contribute to the Wiki? please see some beginners instructions at the [[WikiGetStarted]] page. | ||
Latest revision as of 02:55, 21 September 2016
Welcome To BioDAS.org
Contents
Notice
PLEASE NOTE: a new version of DAS was recently released! See the Versions of DAS section below for more information.
About DAS
The Distributed Annotation System (DAS) defines a communication protocol used to exchange annotations on genomic or protein sequences. It is motivated by the idea that such annotations should not be provided by single centralized databases, but should instead be spread over multiple sites. Data distribution, performed by DAS servers, is separated from visualization, which is done by DAS clients. The advantages of this system are that control over the data is retained by data providers, data is freed from the constraints of specific organisations and the normal issues of release cycles, API updates and data duplication are avoided.
DAS is a client-server system in which a single client integrates information from multiple servers. It allows a single machine to gather up sequence annotation information from multiple distant web sites, collate the information, and display it to the user in a single view. Little coordination is needed among the various information providers.
DAS is heavily used in the genome bioinformatics community. Over the last years we have also seen growing acceptance in the protein sequence and structure communities.
What can I do with DAS ?
A DAS-enabled website or application can aggregate complex and high-volume data from external providers in an efficient manner. For the biologist, this means the ability to "plug in" the latest data, possibly including a user's own data. For the application developer, this means protection from data format changes and the ability to add new data with minimal development cost.
Here are some examples of DAS-enabled applications or websites for end users:
- Dalliance Interactive web-based genome viewer
- IGV Integrative Genome Viewer java based browser for many genomes
- Ensembl uses DAS to pull in genomic, gene and protein annotations. It also provides data via DAS.
- Gbrowse is a generic genome browser, and is both a consumer and provider of DAS.
- IGB is a desktop application for viewing genomic data.
- SPICE is an application for projecting protein annotations onto 3D structures.
- Dasty2 is a web-based viewer for protein annotations
- Jalview is a multiple alignment editor.
- PeppeR is a graphical viewer for 3D electron microscopy data.
- DASMI is an integration portal for protein interaction data.
- DASher is a Java-based viewer for protein annotations.
- EpiC presents structure-function summaries for antibody design.
- STRAP is a STRucture-based sequence Alignment Program.
Hundreds of DAS servers are currently running worldwide, including those provided by the European Bioinformatics Institute, Ensembl, the Sanger Institute, UCSC, WormBase, FlyBase, TIGR, and UniProt. For a listing of all available DAS sources please visit the DasRegistry.
Versions of DAS
The original DAS specification was written by Lincoln Stein, Sean Eddy, and Robin Dowell. It is widely adopted and well supported, particularly throughout Europe, and is the basis for a large number of existing clients and servers. The protocol has been developed incrementally since its inception and thus there are several successive versions of the specification, each expanding on the last whilst retaining a focus on backwards compatibility. The current specification of DAS is version 1.6.
Though mature, the protocol continues to be extended to cater for the needs of the DAS community via extensions to the specification. Together, these extensions form an "extended specification". The current version is 1.6E.
NOTE: The DAS/2 project is an entirely separate specification which although based on the DAS architecture is not backwards compatible with existing servers and clients. It is described separately on the DAS/2 page.
The various specification versions on this site are described here.
Evolution
Graphic representation of the evolution of "Versions of DAS" over the years. The different colors identify the version/specification and the white numbers an estimation of available sources per year.
Further Reading
If you would like to find out more, see a description of the DAS protocol. Additional information is also available in the form of an FAQ page, and a more extensive reference: Everything DAS. There is also a list of DAS related publications.
The Community Portal page contains information on the various DAS mailing lists.
DAS is often represented in workshops, training courses and conferences. A list of past and future events is available on the Current events page.
Contribute to this Wiki
Want to contribute to the Wiki? please see some beginners instructions at the WikiGetStarted page.