Difference between revisions of "DAS/2"
m (Reverted edits by Irywezijyzi (Talk) to last revision by DavidNix) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
== About DAS/2 == | == About DAS/2 == | ||
| − | DAS/2 is intended to be a more flexible and powerful version of the original | + | DAS/2 is intended to be a more flexible and powerful version of the original [[DAS/1]] specification. It was built to address the needs of distributing massive genomic data sets derived from high density microarray applications and Next (and Next Next) Generation Sequencing. Unlike DAS/1, DAS/2 does not require data exchange through text based XML but allows for data distribution using any text or binary format. The two versions are not natively compatible. The DAS/2 code base consists of: |
# DAS/2 Specification: | # DAS/2 Specification: | ||
| Line 25: | Line 25: | ||
Given that the DAS/2 specification is not backward compatible with the DAS/1 version, existing DAS software will continue to support DAS/1. A [http://lists.open-bio.org/pipermail/das2/2008-October/001055.html DAS proxy server is in development] that will permit a DAS/1 server to be accessed by DAS/2 clients. | Given that the DAS/2 specification is not backward compatible with the DAS/1 version, existing DAS software will continue to support DAS/1. A [http://lists.open-bio.org/pipermail/das2/2008-October/001055.html DAS proxy server is in development] that will permit a DAS/1 server to be accessed by DAS/2 clients. | ||
| − | |||
| − | |||
== DAS/2 Clients == | == DAS/2 Clients == | ||
| Line 33: | Line 31: | ||
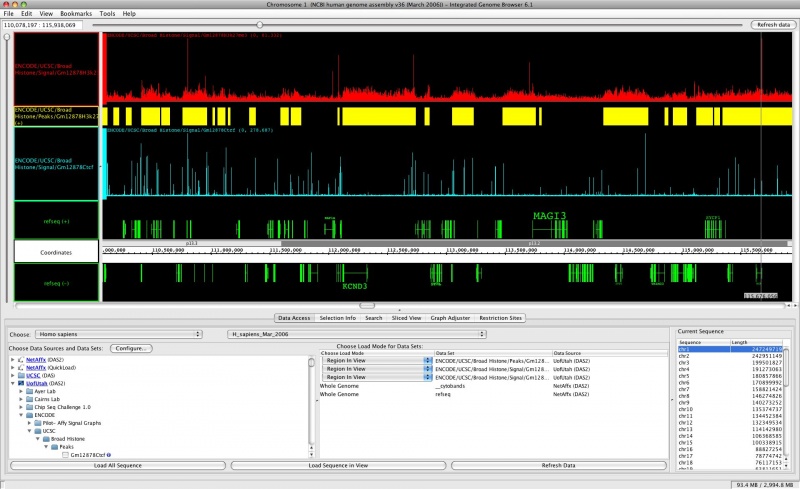
* [http://genoviz.sourceforge.net IGB - Integrated Genome Browser] - a DAS/2 reference implementation | * [http://genoviz.sourceforge.net IGB - Integrated Genome Browser] - a DAS/2 reference implementation | ||
| + | [[File:Igb.jpg | 800px]] | ||
* [http://www.gmod.org/GBrowse GBrowse - Generic Genome Browser] | * [http://www.gmod.org/GBrowse GBrowse - Generic Genome Browser] | ||
* [http://cabio.nci.nih.gov/NCICB/infrastructure/cacore_overview/caBIO caBIO Infrastructure Objects] ''(confirm this)'' | * [http://cabio.nci.nih.gov/NCICB/infrastructure/cacore_overview/caBIO caBIO Infrastructure Objects] ''(confirm this)'' | ||
| Line 44: | Line 43: | ||
* [http://netaffxdas.affymetrix.com/das2 Affymetrix Public DAS/2 server] | * [http://netaffxdas.affymetrix.com/das2 Affymetrix Public DAS/2 server] | ||
* [http://bioserver.hci.utah.edu:8080/DAS2DB/ Huntsman Cancer Institute's DAS/2 server] | * [http://bioserver.hci.utah.edu:8080/DAS2DB/ Huntsman Cancer Institute's DAS/2 server] | ||
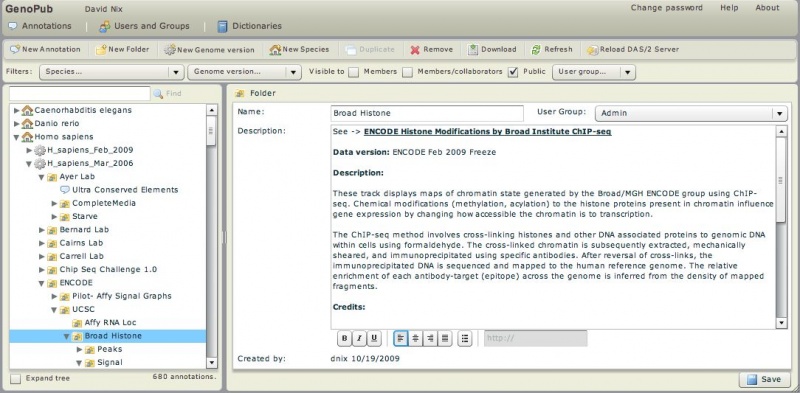
| − | * [http://bioserver.hci.utah.edu:8080/DAS2DB/genopub HCI's GenoPub DAS/2 GUI] - A Flex/MySQL web application for organizing, annotating, and authorizing genomic annotations for distribution using the GenoViz DAS/2 server | + | * [http://bioserver.hci.utah.edu:8080/DAS2DB/genopub HCI's GenoPub DAS/2 GUI] ('''login''': guest, '''pw''': guest) - A Flex/MySQL web application for organizing, annotating, and authorizing genomic annotations for distribution using the GenoViz DAS/2 server |
| − | [[File:Genopub.jpg]] | + | [[File:Genopub.jpg | 800px]] |
* [http://das.biopackages.net/das/genome Biopackages DAS/2 server] - (inactive?) a DAS/2 reference implementation | * [http://das.biopackages.net/das/genome Biopackages DAS/2 server] - (inactive?) a DAS/2 reference implementation | ||
| Line 71: | Line 70: | ||
== Feedback and Bug Reports == | == Feedback and Bug Reports == | ||
| − | A general forum for advice on DAS/2-related issues is the [http://biodas.org/mailman/listinfo/ | + | A general forum for advice on DAS/2-related issues is the [http://biodas.org/mailman/listinfo/das discussion list]. |
To submit or view bug reports in DAS/2-related software, use one of the links below, depending on the affected component. | To submit or view bug reports in DAS/2-related software, use one of the links below, depending on the affected component. | ||
| Line 79: | Line 78: | ||
* Reference Server (GMOD Project - select category 'DAS2'): | * Reference Server (GMOD Project - select category 'DAS2'): | ||
** http://sourceforge.net/tracker/?group_id=27707&atid=391291 | ** http://sourceforge.net/tracker/?group_id=27707&atid=391291 | ||
| − | * Reference Client (IGB): | + | * Reference Client (IGB) - select category 'DAS': |
** http://sourceforge.net/tracker/?group_id=129420&atid=714744 | ** http://sourceforge.net/tracker/?group_id=129420&atid=714744 | ||
* Validation Suite (Dasypus): | * Validation Suite (Dasypus): | ||
** https://sourceforge.net/tracker/?group_id=138271&atid=740641 | ** https://sourceforge.net/tracker/?group_id=138271&atid=740641 | ||
| − | |||
| − | |||
== CVS Access == | == CVS Access == | ||
Latest revision as of 16:17, 16 November 2010
The DAS/2 Protocol
Contents
About DAS/2
DAS/2 is intended to be a more flexible and powerful version of the original DAS/1 specification. It was built to address the needs of distributing massive genomic data sets derived from high density microarray applications and Next (and Next Next) Generation Sequencing. Unlike DAS/1, DAS/2 does not require data exchange through text based XML but allows for data distribution using any text or binary format. The two versions are not natively compatible. The DAS/2 code base consists of:
- DAS/2 Specification:
- DAS/2.0 specification (stable)
- DAS/2.1 specification (under development)
- (Inactive ?) A publicly accessible DAS/2 server implementation (open source code available from the GMOD project)
- The GenoViz open-source DAS/2 client implementation
- An open-source client library and server validation suite
See DAS/2#CVS Access for details on retrieving code related to these projects.
DAS/2 History
DAS/2 diverged from DAS/1 as a result of a variety of RFCs (Requests for Comment) posted on the DAS mailing list from users and implementers of the DAS spec over the first several years after DAS was originally introduced. As of November 2006, the retrieval portion of the DAS/2 spec has stabilized. Implementers and service providers can rely on this for their DAS/2-based development efforts.
Work on what became the DAS/2.0 specification "officially" began in July 2004 when funding was awarded for a 2-year NIH grant for DAS/2 development (abstract, full proposal). The grant was subsequently extended, and the grant funding period ended October 2007 (final progress report). Participating in the DAS/2 grant were Affymetrix, Cold Spring Harbor Lab, the European Bioinformatics Institute/Sanger Center, and Dalke Scientific.
Given that the DAS/2 specification is not backward compatible with the DAS/1 version, existing DAS software will continue to support DAS/1. A DAS proxy server is in development that will permit a DAS/1 server to be accessed by DAS/2 clients.
DAS/2 Clients
The following software packages support the client-side DAS/2 API for interacting with DAS/2-compliant servers:
- IGB - Integrated Genome Browser - a DAS/2 reference implementation
- GBrowse - Generic Genome Browser
- caBIO Infrastructure Objects (confirm this)
DAS/2 Servers
The following software packages operate as servers capable of providing data in response to queries conforming with the DAS/2 protocol:
- Affymetrix Public DAS/2 server
- Huntsman Cancer Institute's DAS/2 server
- HCI's GenoPub DAS/2 GUI (login: guest, pw: guest) - A Flex/MySQL web application for organizing, annotating, and authorizing genomic annotations for distribution using the GenoViz DAS/2 server
- Biopackages DAS/2 server - (inactive?) a DAS/2 reference implementation
DAS/2 Validation Suite
See Das2Validation.
Publishing, Discovering, and Registering DAS/2 sources
Currently, the DasRegistry does not support DAS/2 services.
As a temporary fix, download the 'das2Registry.xml' file from the GenoViz Project, add an entry, and email it back to GenoViz or BioDas. If you have access to the GenoViz SVN, modify it directly.
Global Sequence Identifiers
See GlobalSeqIDs
Feedback and Bug Reports
A general forum for advice on DAS/2-related issues is the discussion list.
To submit or view bug reports in DAS/2-related software, use one of the links below, depending on the affected component.
- DAS/2 Specification:
- Reference Server (GMOD Project - select category 'DAS2'):
- Reference Client (IGB) - select category 'DAS':
- Validation Suite (Dasypus):
CVS Access
There are separate repositories for the DAS/2 specification, DAS/2 client and server reference implementations, and the validation suite. These are for developers only since they do not represent stable releases and may contain modifications that contain bugs. The one exception is the schema for the retrieval portion of the DAS/2 spec, which is now stable.
- DAS/2 spec:
-
:pserver:cvs@cvs.biodas.org:/home/repository/biodas- (login password="cvs", directory=das/das2, XML schema for retrieval spec: das2_schemas.rnc)
-
- DAS/2 Client reference implementation ("IGB")
- DAS/2 Server reference implementation (see das2 package)
- DAS/2 validation suite ("Dasypus")
See http://www.open-bio.org/wiki/SourceCode for general pointers about anonymous access to open-bio.org-hosted source code.