| Name | Type | Use | Default | Fixed | Annotation | ||
| ls:lsid | lsid | ||||||
| tns:length | integer | ||||||
| number | string | optional |
|
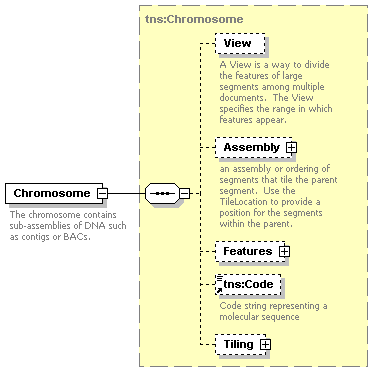
| documentation | The chromosome contains sub-assemblies of DNA such as contigs or BACs. |
| schema location: | C:\Dev\omnigene\resources\das\annotation.xsd |
| targetNamespace: | http://www.biodas.org/names/annotation |
| Elements | Complex types |
| Chromosome | Chromosome |
| Code | Contig |
| Contig | DataSource |
| Feature | Feature |
| Location | Location |
| Segment | Segment |
| TileLocation | TileLocation |
| Type | Type |
| schema location: | C:\Dev\omnigene\resources\das\lscore.xsd |
| targetNamespace: | http://www.i3c.org/names/core |
| Elements | Simple types |
| lsid | lsid |
| diagram |  |
||||||||||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||||||||||
| type | tns:Chromosome | ||||||||||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||
| annotation |
|
| diagram |  |
||
| namespace | http://www.biodas.org/names/annotation | ||
| type | string | ||
| used by |
|
||
| annotation |
|
| diagram | 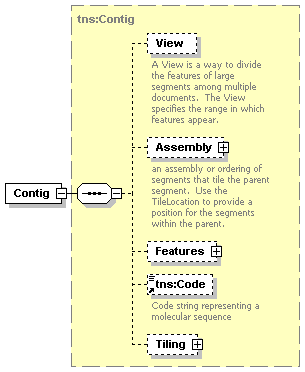 |
||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| type | tns:Contig | ||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
| diagram | 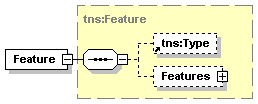 |
||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||
| type | tns:Feature | ||||||||||||
| children | tns:Type Features | ||||||||||||
| used by |
|
||||||||||||
| attributes |
|
| diagram | |||||||||||||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||||||||||||
| type | tns:Location | ||||||||||||||||||||||||||||
| attributes |
|
| diagram | 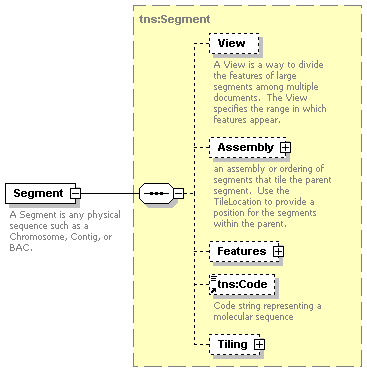 |
||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| type | tns:Segment | ||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||
| attributes |
|
||||||||||||||||||
| annotation |
|
| diagram |  |
||||||||||||||||||||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||||||||||||||||||||
| type | tns:TileLocation | ||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||
| annotation |
|
| diagram | |||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| type | tns:Type | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
| diagram | 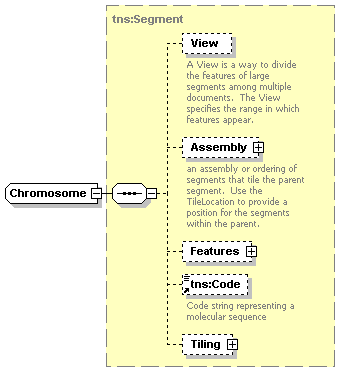 |
||||||||||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||||||||||
| type | extension of tns:Segment | ||||||||||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||
| attributes |
|
| diagram | 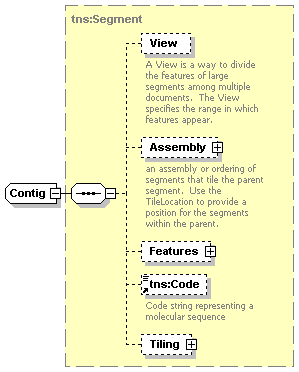 |
||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| type | extension of tns:Segment | ||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
| diagram | |||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||
| children | description | ||||||||||||
| attributes |
|
| diagram | |
| type | string |
| diagram | 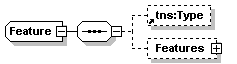 |
||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||
| children | tns:Type Features | ||||||||||||
| used by |
|
||||||||||||
| attributes |
|
| diagram | |
| children | tns:Feature |
| diagram |  |
||||
| namespace | http://www.biodas.org/names/annotation | ||||
| used by |
|
||||
| attributes | |||||
| annotation |
|
| diagram | 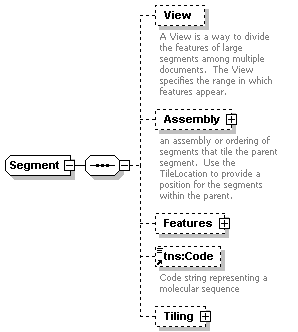 |
||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| children | View Assembly Features tns:Code Tiling | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
| diagram |  |
||||||||||||||||||||||||||||
| type | extension of tns:Location | ||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||
| annotation |
|
| diagram |  |
||
| children | tns:Contig | ||
| annotation |
|
| diagram | |
| children | tns:Feature |
| diagram |  |
| children | tns:TileLocation |
| diagram | |||||||||||||||||||||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||||||||||||||||||||
| type | extension of tns:Location | ||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||
| attributes |
|
| diagram | |||||||||||||||||||
| namespace | http://www.biodas.org/names/annotation | ||||||||||||||||||
| used by |
|
||||||||||||||||||
| attributes |
|
| diagram |  |
||||||||||||
| namespace | http://www.i3c.org/names/core | ||||||||||||
| attributes |
|
||||||||||||
| annotation |
|
| namespace | http://www.i3c.org/names/core | ||
| type | restriction of string | ||
| used by |
|
||
| facets |
|
||
| annotation |
|